A. Growth dynamics of C. elegans cuticle mutants
Characterization of larval growth in C. elegans cuticle mutants
Joy Nyaanga, Sasha Shirman, Niall M. Mangan, and Erik C. Andersen
Abstract
In Caenorhabditis elegans, many genes involved in the formation of the cuticle are also known to influence body size and shape. We assessed post-embryonic growth of both long and short C. elegans body size mutants from the L1 to L4 stage. We found similar developmental trajectories of N2 and lon-3 animals. By contrast, we observed overall decreases in body length and increases in body width of tested dpy mutants compared to N2, consistent with the Dpy phenotype. We further show that the dynamics of animal shape in the mutant strains are consistent with a previously proposed “Stretcher” growth model.
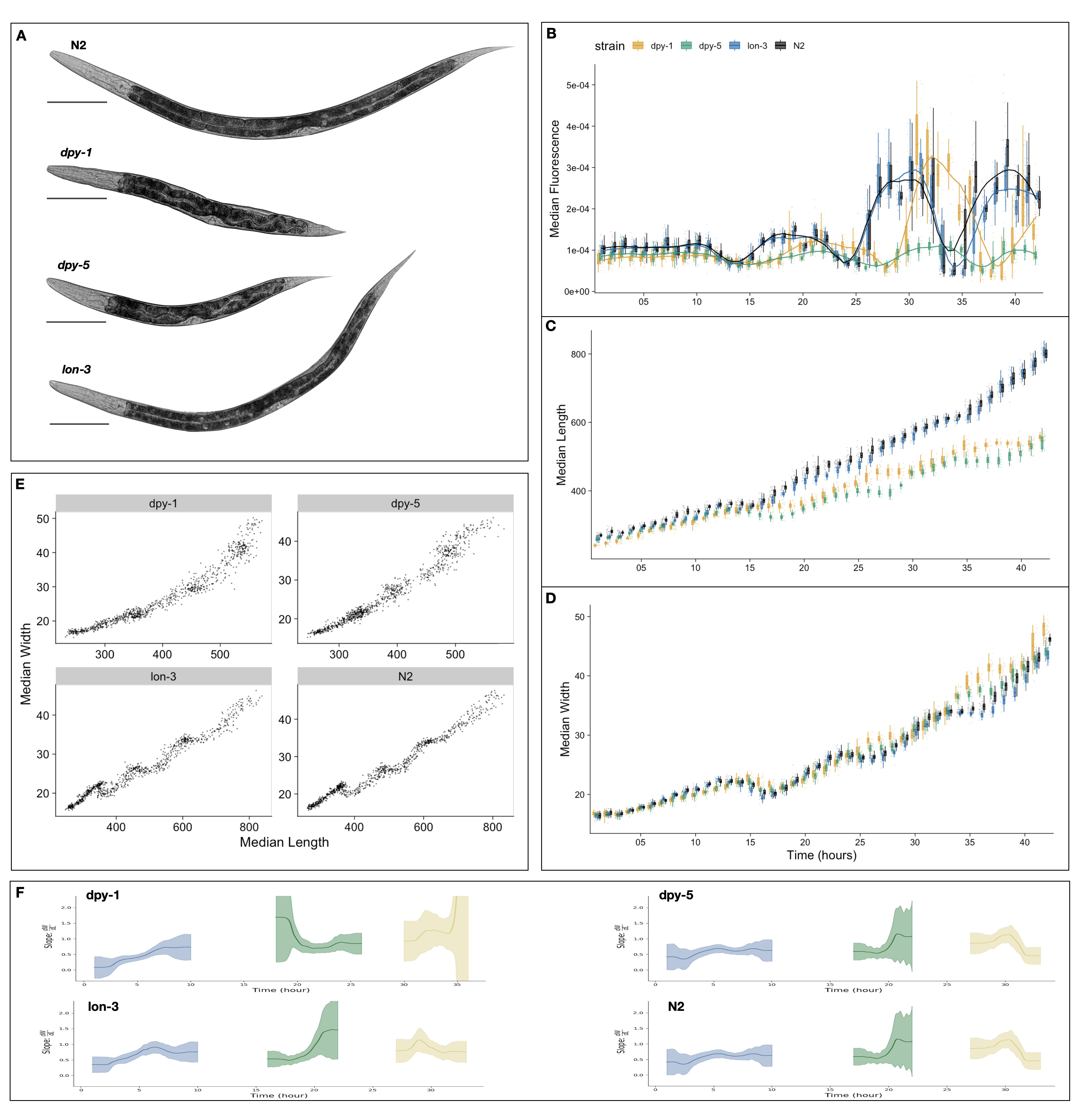
Figure A-1: Quantitative assessment of C. elegans larval growth. (A) Representative images of strains used in this study taken at the L4 stage. Tukey boxplots for median animal fluorescence normalized by area (B), median animal length (\(\mu M\)) (C), and median animal width (\(\mu M\)) (D). The horizontal line in the middle of the box is the median, and the box denotes the 25th to 75th quantiles of the data. The vertical line represents the 1.5 interquartile range. Each point corresponds to the median value of a population of animals in each well. (E) Median length (x-axis) plotted against median length (y-axis). (F) The ratio of the change in width to length over time. Calculated from the local slope of data in panel (E). The standard deviation captures population variation (grey).
Description
Body size largely influences an organism’s functional characteristics: growth, reproduction, metabolism, lifespan. As a result, the determinant factors of organism size, particularly during development, have been explored in many systems. Caenorhabditis elegans, a free-living nematode, presents a versatile genetic model system to study how the processes of growth and development are regulated. C. elegans matures to an adult after multiple molting events during which time animals synthesize a new exoskeleton (cuticle) and expel their old one. The C. elegans cuticle is a complex, multi-layered structure primarily composed of collagens. As animals progress through their life-cycle, the structure and thickness of the cuticle changes but its role in the maintenance of body morphology and integrity remains. To date, 21 cuticle collagen mutants have been identified that cause a range of body morphology defects (Page and Johnstone 2007). Some of these mutants exhibit a disproportionate reduction in body size, while others are noticeably larger than wild type (Cho et al. 2021), clearly demonstrating the importance of the physical structure of the cuticle on growth. Analyzing the characteristics of size during development in various C. elegans body shape mutants is central to understanding the role these genetic pathways have on body growth.
We performed a high-resolution longitudinal study of growth in a selection of C. elegans cuticle mutants. We selected mutants that were both reportedly shorter (dpy-1(e1), dpy-5(e61)) and longer (lon-3(e2175)) than wild type (Cho et al. 2021) (Figure A-1A). We then collected high-precision measurements of animal fluorescence (Figure A-1B), length (Figure A-1C), and width (Figure A-1D) from the L1 stage through the L4 stage. As we previously demonstrated (Nyaanga et al. 2022) we can use oscillations in fluorescence as a proxy for feeding behavior to characterize larval progression by associating periods of decreased feeding with molt events. Doing so, we notice that lon-3(e2175) animals follow similar developmental trajectories to the N2 wild type. By contrast, we detect a delay in the molt timing of both dpy mutant strains, with dpy-1(e1) undergoing each larval transition later than all other tested strains. We also observe a marked decrease in animal length and increase in animal width noticeable after the L1 stage in both dpy strains, consistent with their characteristic dumpy phenotype. Interestingly, we note little size divergence between lon-3(e2175) and N2 animals during our time course.
Measurements of both animal length and width allow us to assess changes in body shape as well as size. Previously, motivated by changes in the body aspect ratio of animals we observed at larval stage transitions, we modeled a physical mechanism by which constraints on cuticle stretch could cause changes in C. elegans body shape (Nyaanga et al. 2022). We found that model-predicted shape changes were consistent with those seen in our data of N2 animals. Given this result, we proposed a “Stretcher” model for growth wherein C. elegans sense changes in cuticle elasticity, in tandem with other regulatory mechanisms, to control growth rate and determine developmental transitions. Given the structural impacts of dpy-1(e1), dpy-5(e61) and lon-3(e2175) mutants, we sought to determine whether the shape dynamics predicted by the Stretcher model would be consistent with the mutant data. By analyzing the relationship between measured animal length and width (W/L) over time, we are able to detect the linear and nonlinear stretch regimes predicted by the Stretcher model (Figure A-1F). In all strains, we observe an approximately constant W/L ratio in all larval stages, consistent with a linear stretch regime. Next, we observe a shape slope increase, consistent with a nonlinear stretch regime in length preceding larval stage transitions.
Methods
Worm culture
The laboratory strain N2 was obtained from the C. elegans Natural Diversity Resource (Cook et al. 2017). All other strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Animals were cultured at 20C on 6 cm plates of modified nematode growth media (NGMA), which contained 1% agar and 0.7% agarose seeded with E. coli OP50 bacteria.
High-throughput growth assay
Measurements of body size and fluorescence were measured as previously described (Nyaanga et al. 2022). Briefly, strains were propagated for three generations, bleach-synchronized, and titered at a concentration of 1 embryo per \(\mu L\) into 250 mL flasks. The following day, arrested L1s were fed HB101 food at a final concentration of OD20 in a final flask volume of 100 mL K medium and HB101 food. Animals were grown with constant shaking at 20C. Flasks were sampled each hour beginning one hour after feeding and continuing for 42 consecutive hours. At each hour, 800 \(\mu L\) was removed from each flask and incubated with fluorescent polychromatic beads (Polysciences, 19507-5) for 10 minutes with shaking. Following the bead incubation, animals were transferred to a 96-well microtiter plate, treated with sodium azide, imaged with an ImageXpress Nano (Molecular Devices, SanJose, CA), and scored using a large-particle flow cytometer (COPAS BIOSORT, Union Biometrica, Holliston MA). COPAS BIOSORT was used to collect measurements of animal length (TOF), optical extinction (EXT), and red fluorescence for every animal in each well.
Data processing
COPAS BIOSORT data were processed as previously described (Nyaanga et al. 2022). To remove non-animal objects such as bacterial clumps, shed cuticles, and next generation larval animals from the time-course data. Data for each well was summarized to obtain median well measurements. TOF and norm.EXT data were then converted to microns. Only the unit-corrected data were used for further analysis. “Stretcher” model analysis of shape dynamics was performed as previously described (Nyaanga et al. 2022).
Reagents
| STRAIN | GENOTYPE | AVAILABLE FROM |
|---|---|---|
| N2 | Caenorhabditis elegans | CeNDR |
| CB1 | dpy-1(e1) | CGC |
| CB61 | dpy-5(e61) | CGC |
| CB4123 | lon-3(e2175) | CGC |
Contributions
Joy Nyaanga: Conceptualization, Methodology, Investigation, Formal Analysis, Visualization, Writing - original draft
Sasha Shirman: Methodology, Formal Analysis, Visualization
Niall M. Mangan: Methodology, Funding acquisition, Supervision
Erik C. Andersen: Conceptualization, Methodology, Investigation, Funding acquisition, Supervision
References
Page AP, Johnstone IL. The cuticle. WormBook. 2007 Mar 19;1–15. PMCID: PMC4781593
Cho JY, Choi T-W, Kim SH, Ahnn J, Lee S-K. Morphological Characterization of small, dumpy, and long Phenotypes in Caenorhabditis elegans. Mol Cells. Korean Society for Molecular and Cellular Biology; 2021 Mar 31;44(3):160–167. PMCID: PMC8019597
Nyaanga J, Goss C, Zhang G, Ahmed HN, Andersen EJ, Miller IR, Rozenich JK, Swarthout IL, Vaughn JA, Mangan NM, Shirman S, Andersen EC. Changes in body shape implicate cuticle stretch in C. elegans growth control. bioRxiv. 2022. p. 2021.04.01.438121. Available from: https://www.biorxiv.org/content/10.1101/2021.04.01.438121v3
Cook DE, Zdraljevic S, Roberts JP, Andersen EC. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res [Internet]. 2017 Jan 4;45(D1):D650–D657. Available from: http://dx.doi.org/10.1093/nar/gkw893 PMCID: PMC5210618